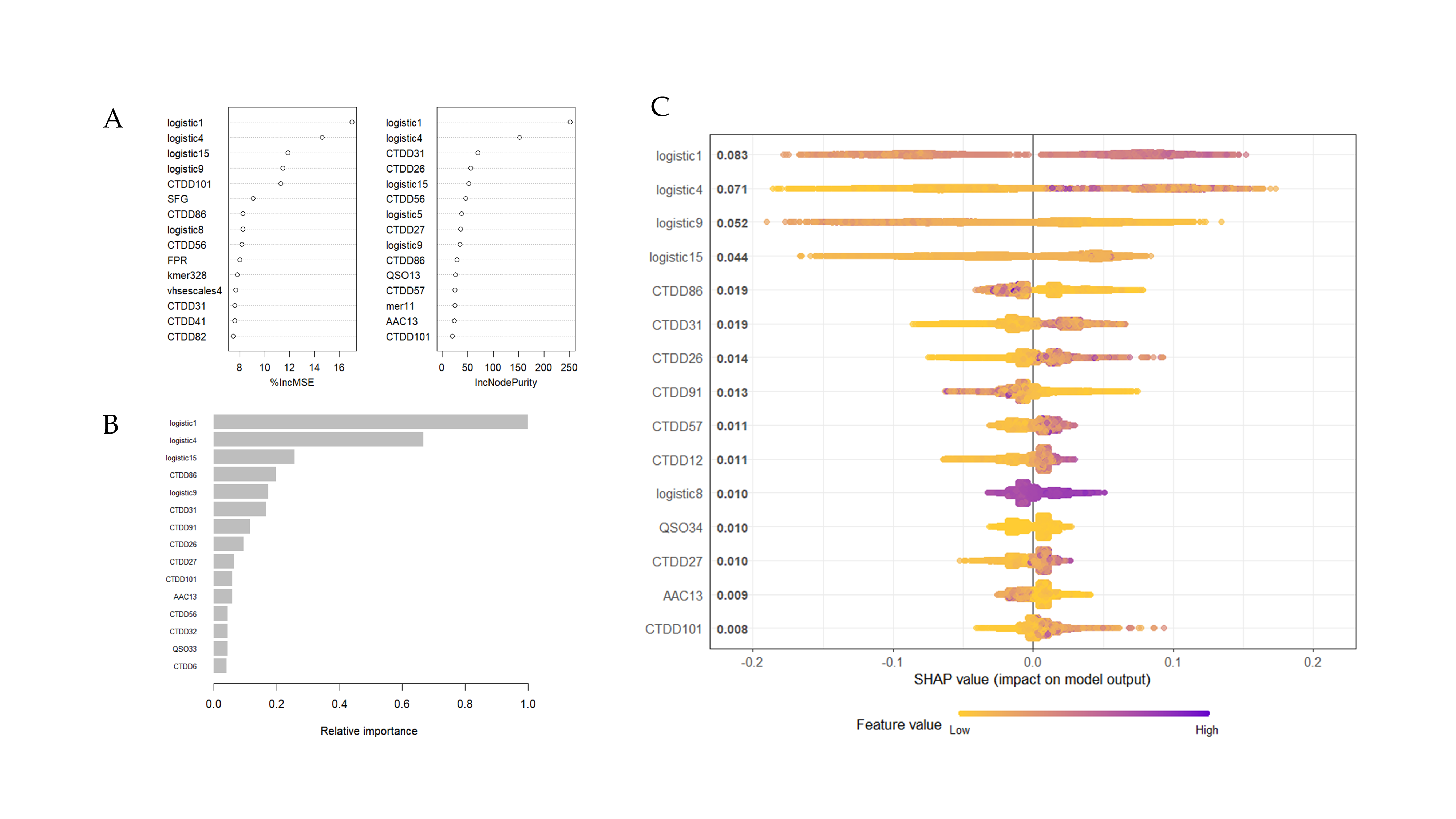
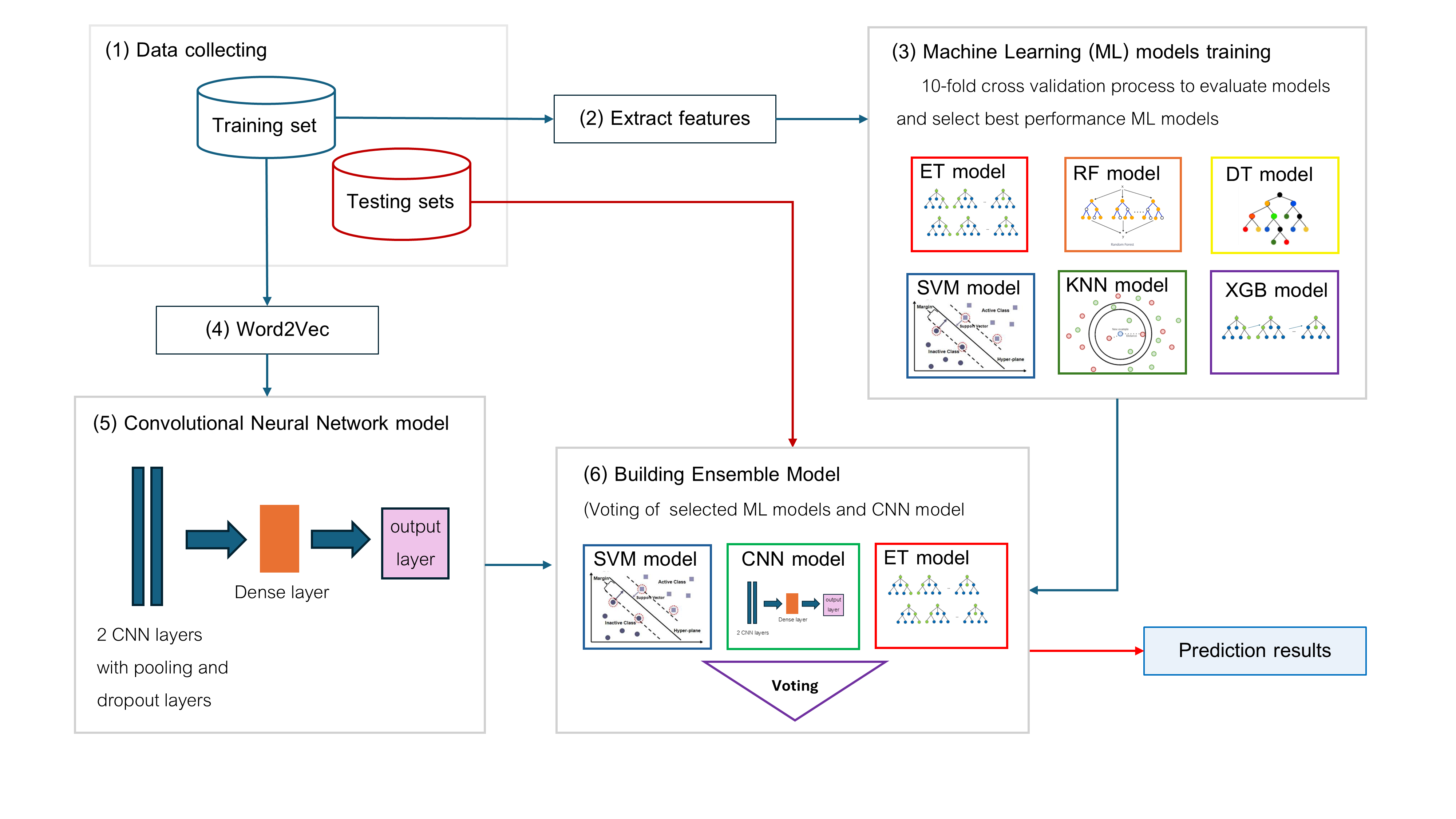
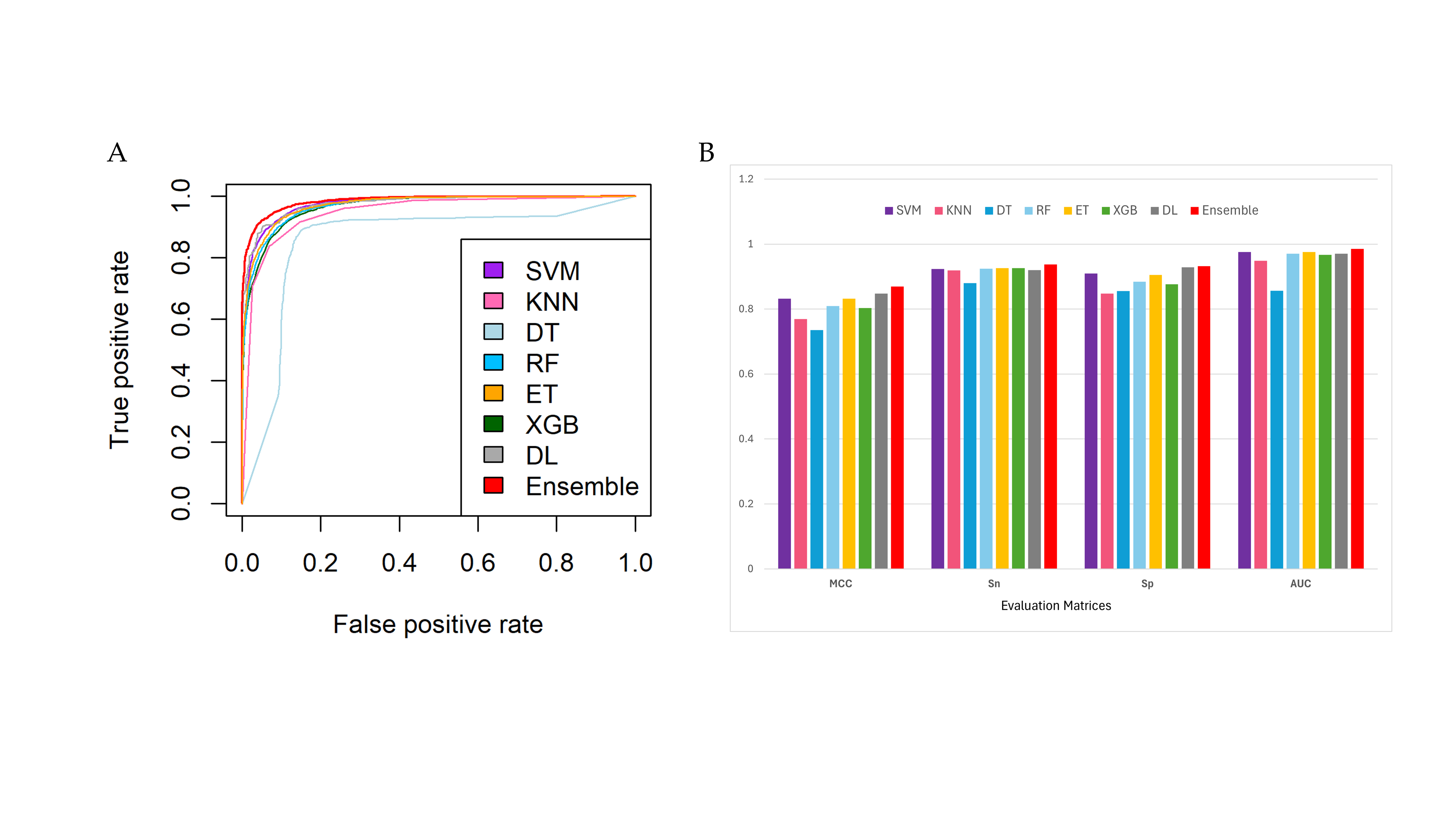
EnsembleNPPred
A
Robust Approach to Neuropeptide Prediction and Recognition Using Ensemble
Learning with Machine learning and Deep learning methods
Download Link:
Training and Testing data, EnsembleNPPred_models
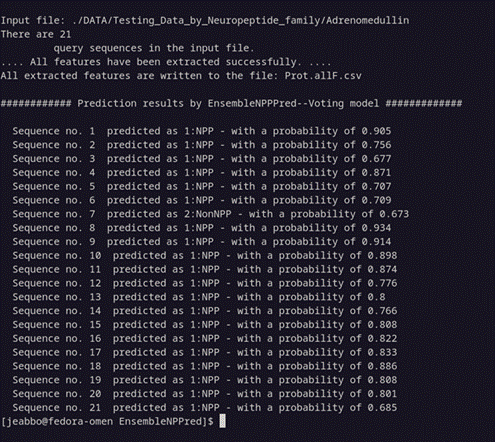
Command: perl EnsembleNPPred.pl input_fasta_file
***** Fasta seq must has length > 5 aa and contains only
standard amino acids *****
###Example:
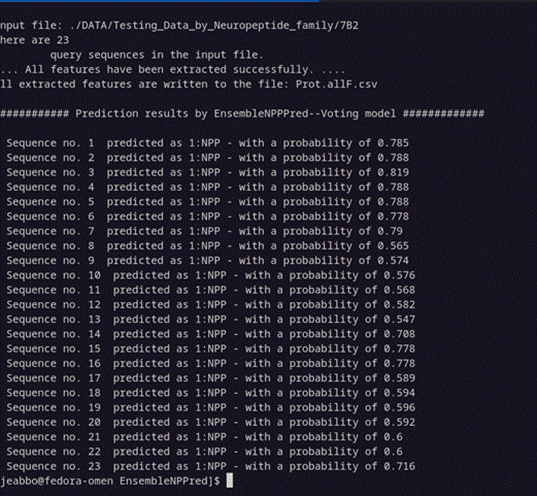
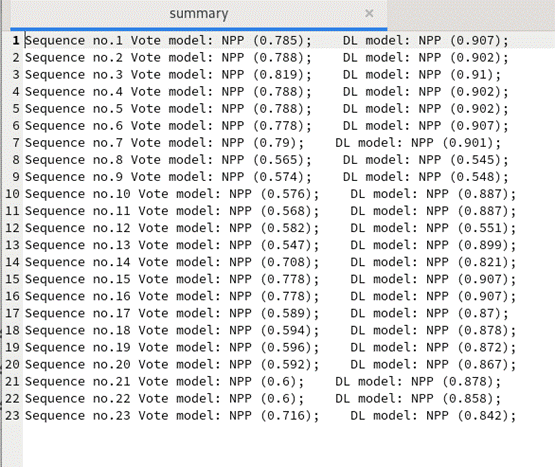
perl EnsembleNPPred.pl
./DATA/Testing_Data_by_Neuropeptide_family/7B2
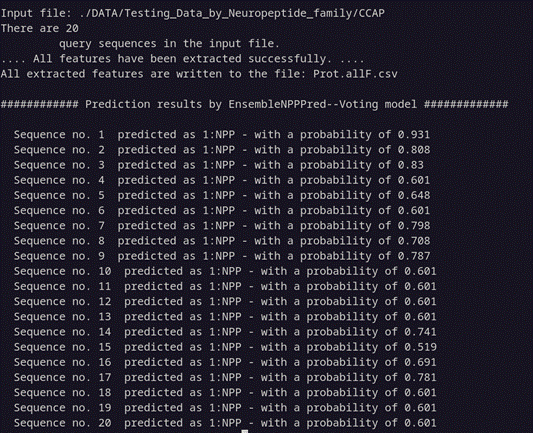
perl EnsembleNPPred.pl ./DATA/Testing_Data_by_Neuropeptide_family/CCAP
perl EnsembleNPPred.pl ./DATA/Testing_Data_by_Neuropeptide_family/Adrenomedullin