Non-coding RNAs identification with Hybrid Random Forest Ensemble algorithm
Abstract
Since
there is a wide spectrum of non-coding RNAs (ncRNAs) from short ncRNAs to long
ncRNAs, it is a great challenge to identify ncRNA signals within genomic
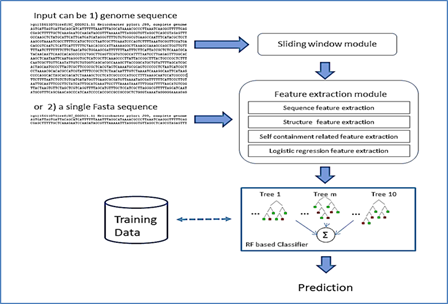
regions. This study has developed a classification tool based on a hybrid
random forest with a logistic regression model to efficiently discriminate not
only those limited to the short ncRNA sequences but also the long and complex
ncRNA classes. This random forest based classifier is
trained on well-balanced training data using a discriminative set of features
and has achieved 92.11%, 90.7% and 93.5% in terms of accuracy, sensitivity and
specificity, respectively. The selected feature set includes a new proposed
feature called SCORE. The SCORE feature is generated based on a logistic
regression function that comprises of five significant features, which
are--structure, sequence, modularity, structural robustness, and coding
potential based features--to better characterize lncRNA functional elements.
This SCORE feature has revealed in this study a useful criterion for finding lncRNAs by improving the performance of the RF-based
classifier in classifying Rfam’s lncRNA families. A
genome-wide ncRNA classification framework has been applied to a wide variety
of organisms especially those potentially important genomes that could have
economic, social, public health, environmental and agricultural impacts such as
various bacteria genomes, Spirulina genome, rice and human genomic
region sequences. The results have reported that our framework is able to
identify known ncRNAs by yielding a sensitivity level above 90% and 77.7% for
prokaryotic and eukaryotic sequences, respectively.
Supplementary Files: Source Code, README, Example files
Identification
of non-coding RNAs with a new composite feature in the Hybrid Random Forest
Ensemble algorithm, Nucleic acids research 42 (11), e93-e93